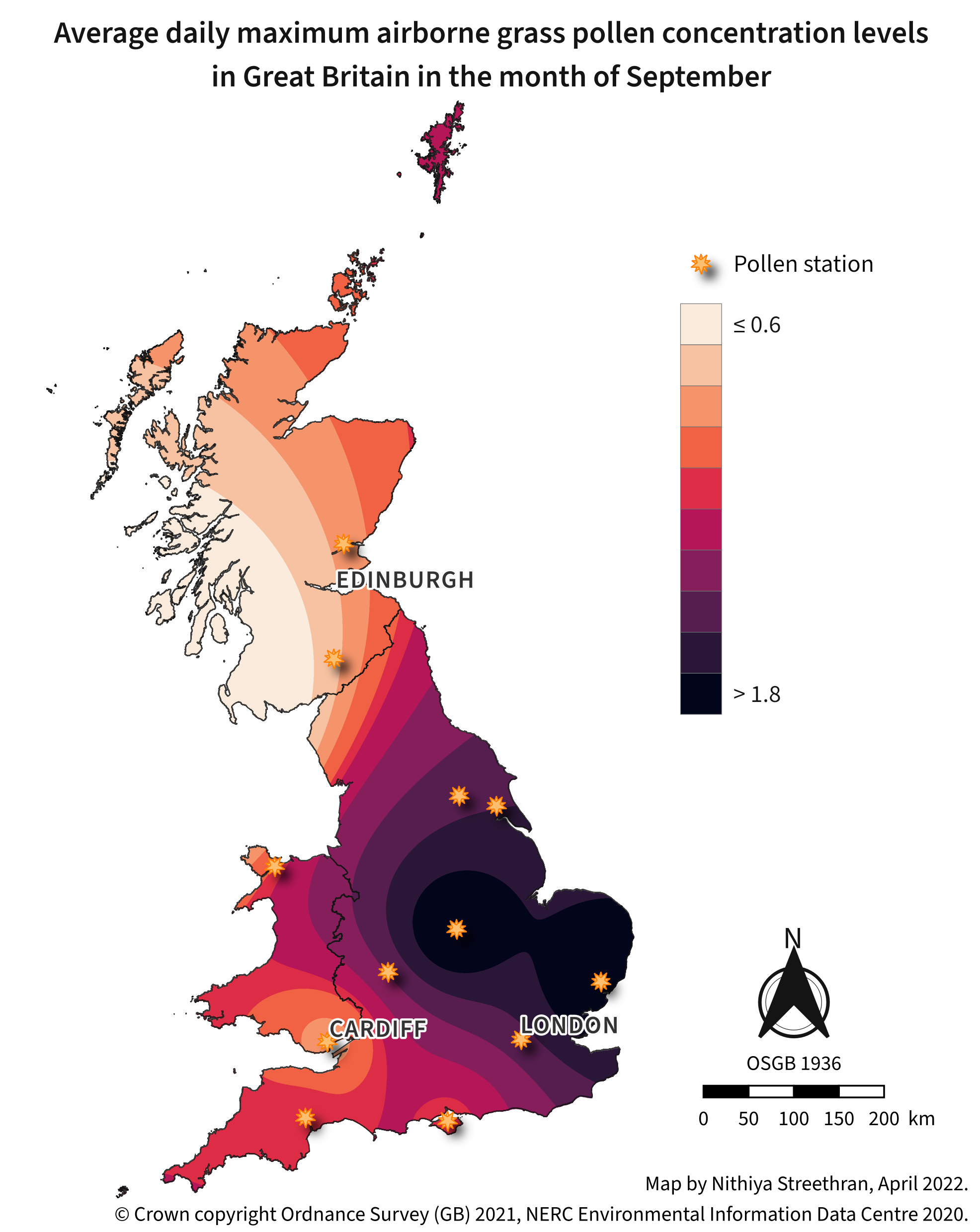
Airborne pollen concentration maps can be a useful indicator of areas to avoid during pollen season for people who suffer from hay fever1.
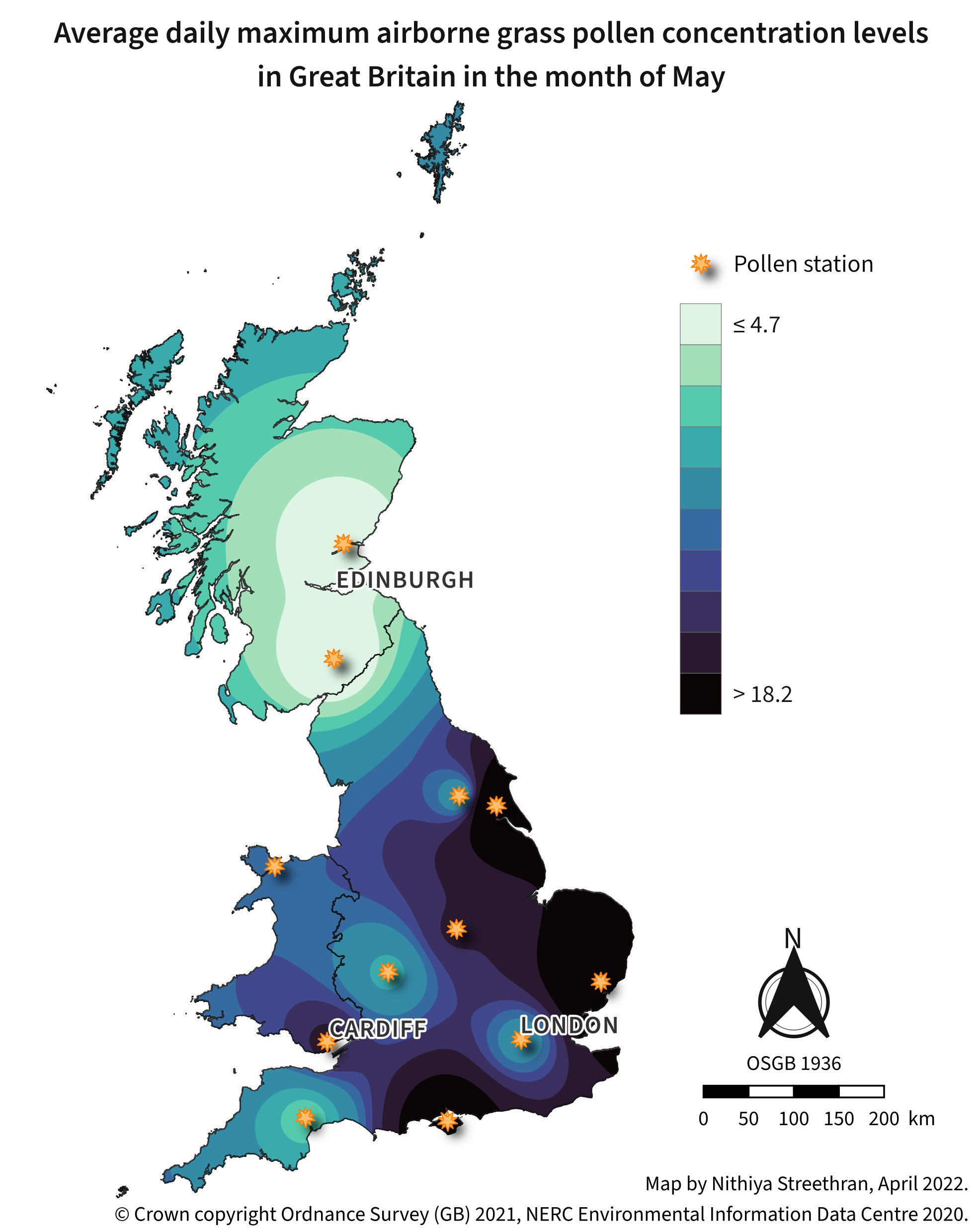
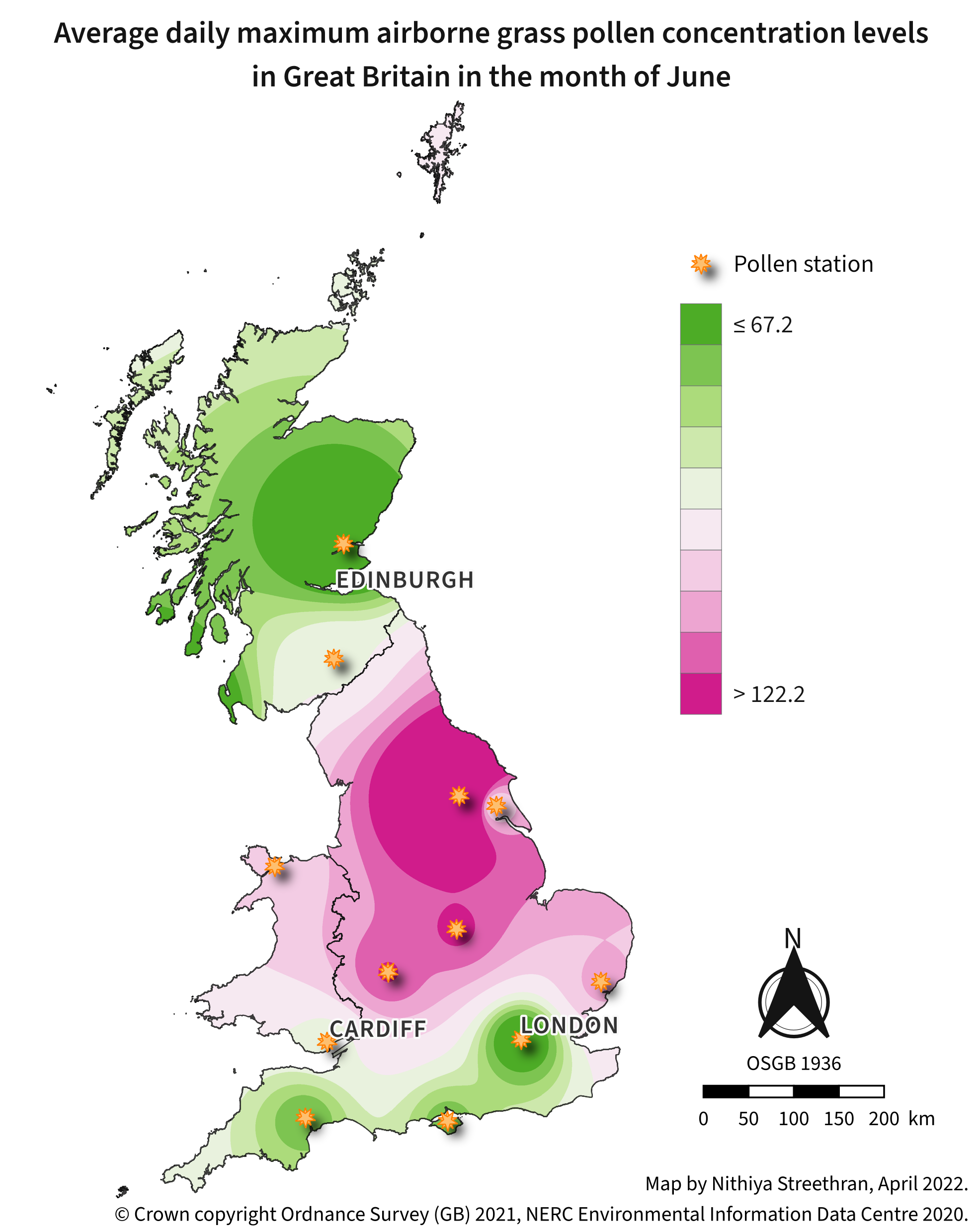
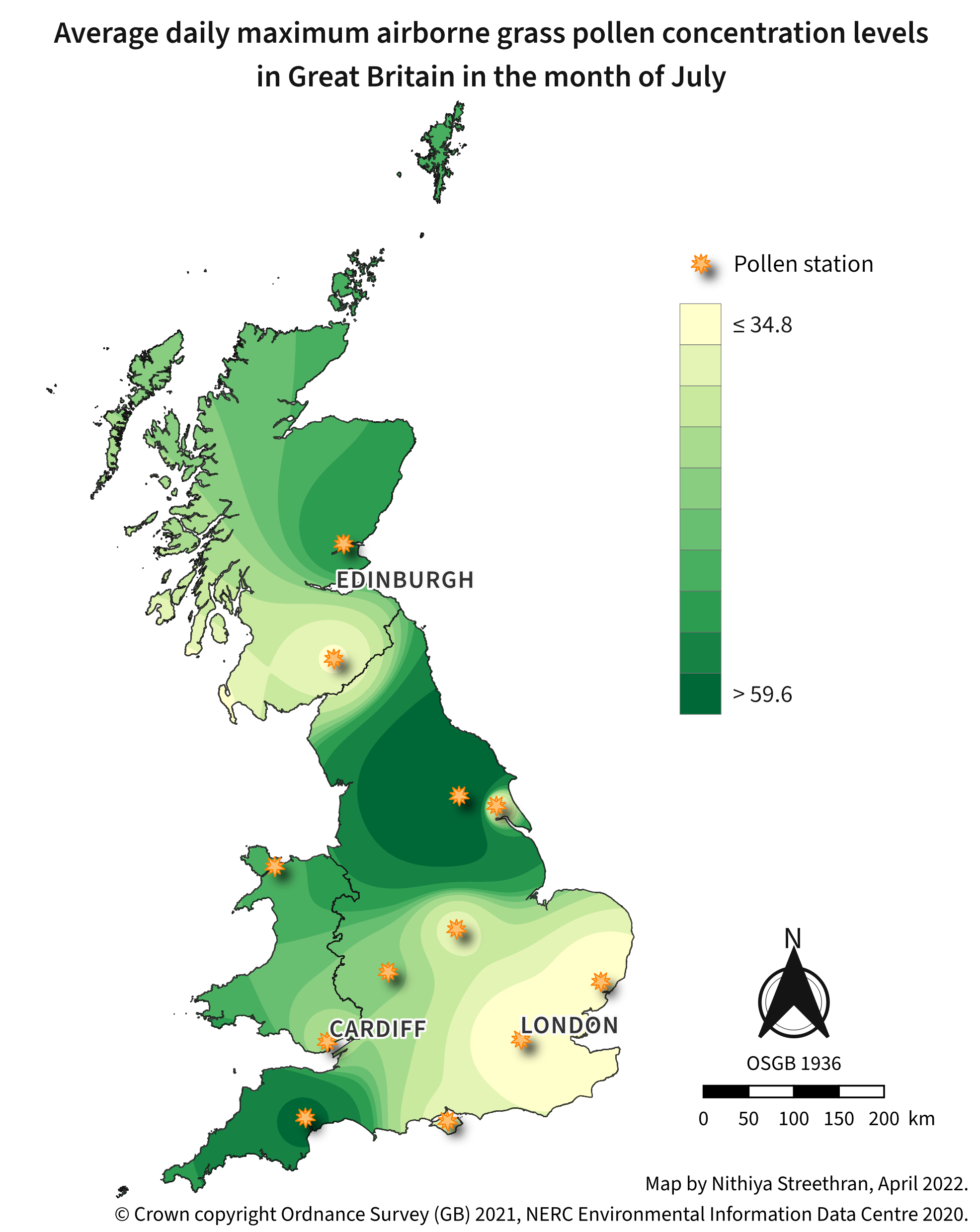
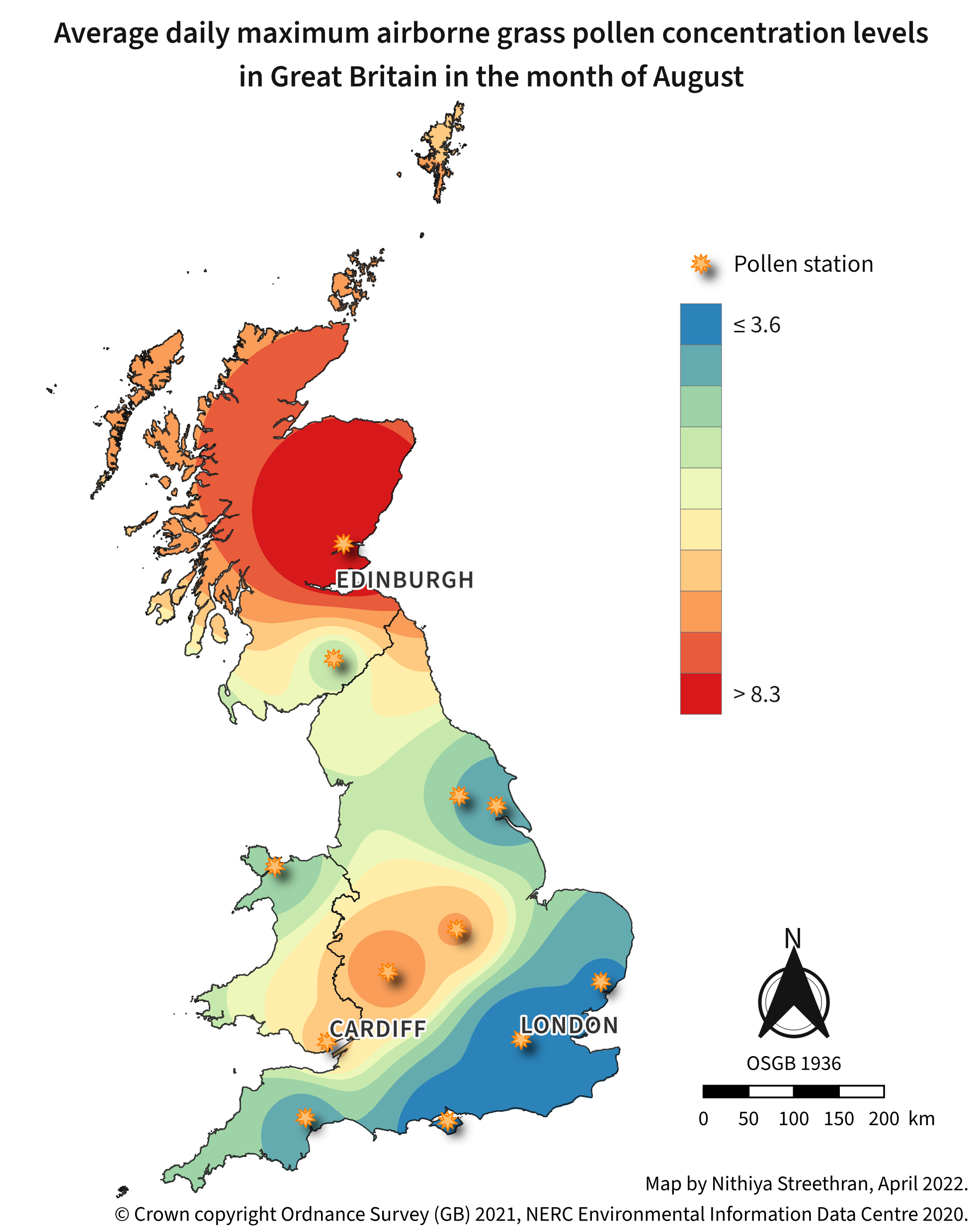
Tools used
Data (licenced under the Open Government Licence, version 3.0 (OGLv3.0)):
- Abundance of airborne pollen for nine grass species, measured by qPCR, UK, 2016-20172
- OS Open Zoomstack, layer
names3 - OS Boundary-Line™, layer
country_region4
Software:
Data processing
First, the pollen data must be restructured into a GeoPackage using the Python script shown in the section below.
The vector point data is interpolated using the Inverse Distance Weighted (IDW) algorithm in QGIS with the following Python snippet:
layerList = ["May", "Jun", "Jul", "Aug", "Sep"]
extent = (
"5512.998000000,655989.000000000,5333.809600000,1220310.000000000" +
" [EPSG:27700]"
)
for layer in layerList:
interpolation_data = (
"data/pollen-data.gpkg|layername=" + layer + "::~::0::~::16::~::0"
)
params = {
"INTERPOLATION_DATA": interpolation_data,
"EXTENT": extent,
"PIXEL_SIZE": 100,
"OUTPUT": "data/idw_" + layer + ".tif"
}
processing.run("qgis:idwinterpolation", params)
Each interpolated raster layer is then clipped to Great Britain’s boundary:
for layer in layerList:
params = {
"INPUT": "data/idw_" + layer + ".tif",
"MASK": "data/bdline_gb.gpkg|layername=country_region",
"SOURCE_CRS": QgsCoordinateReferenceSystem("EPSG:27700"),
"TARGET_CRS": QgsCoordinateReferenceSystem("EPSG:27700"),
"OUTPUT": "data/idw_" + layer + "_clipped.tif"
}
processing.run("gdal:cliprasterbymasklayer", params)
Mapping procedure
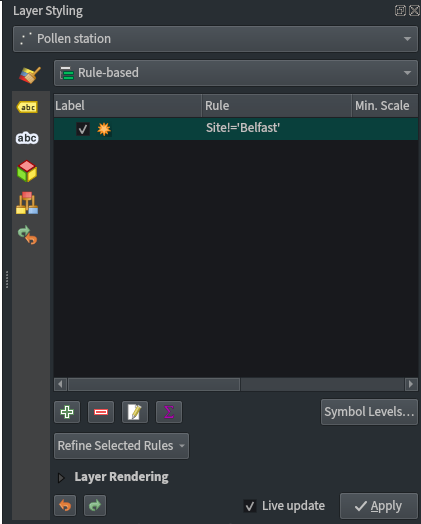
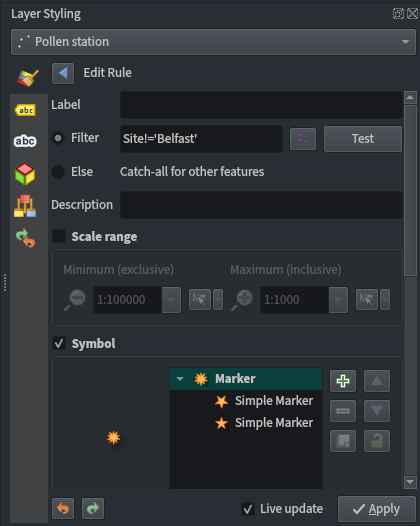
Pollen station point styling:
- Use a rule-based labelling to exclude Belfast using the filter
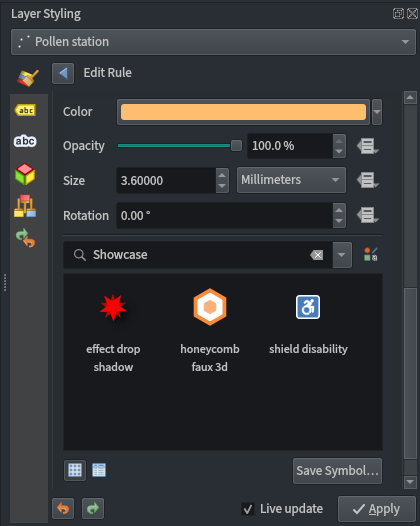
Site!='Belfast' - Use the ‘effect drop shadow’ symbol available in QGIS’ ‘Showcase’ point symbol list
- Change the fill colour to
#fdbf6fand outline to#ff7f00
Basemap styling:
- Use OS stylesheets for the appropriate layers5 6
-
The stylesheets can be applied using the following process:
# Open Zoomstack params = { "INPUT": "data/OS_Open_Zoomstack.gpkg|layername=names", "STYLE": "data/names.qml" } processing.run("native:setlayerstyle", params) # Boundary-Line params = { "INPUT": "data/bdline_gb.gpkg|layername=country_region", "STYLE": "data/country_region.qml" } processing.run("native:setlayerstyle", params)
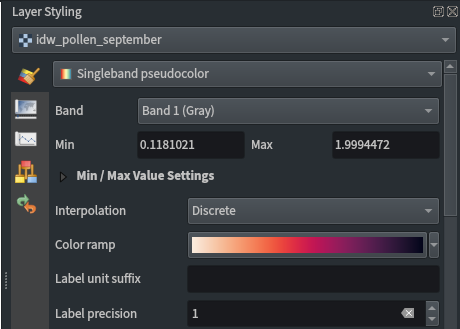
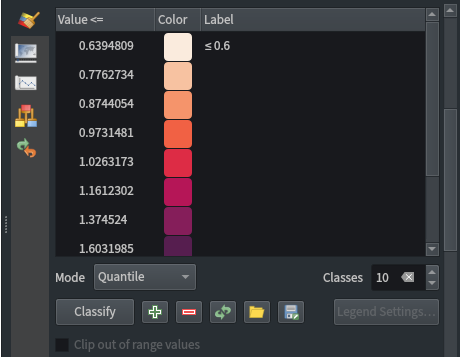
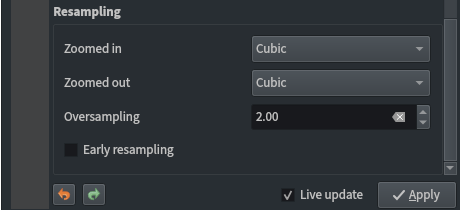
Raster layer styling:
- Singleband pseudocolour
- Discrete interpolation
- Label precision of 1
- Quantile classification mode
- 10 classes
- Cubic resampling
Raster colour ramps:

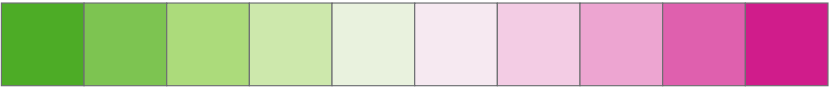



Mako and Rocket are Seaborn colour maps7 8, while the rest are from ColorBrewer9.
Python script to restructure pollen data
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
# import libraries
import pandas as pd
import geopandas as gpd
# read data
pollen = pd.read_csv("data/qPCR_copy_number_abundance_data_aerial_DNA.csv")
# create well known text from coordinates
pollen["wkt"] = (
"POINT (" + pollen["Long"].astype(str) + " " +
pollen["Lat"].astype(str) + ")"
)
# drop cells with no value
pollen = pollen.dropna(subset=["Lat", "Long", "MaxPoaceaeConc", "year-month"])
# use full pollen monitoring site name
pollen = pollen.replace({
"EXE": "Exeter", "EastR": "East Riding", "ESK": "Eskdalemuir",
"LEIC": "Leicester", "CAR": "Cardiff", "IOW": "Isle of Wight",
"IPS": "Ipswich", "BNG": "Bangor", "WOR": "Worcester",
"KCL": "King's College London", "YORK": "York", "ING": "Invergowrie",
"BEL": "Belfast"
})
# create a geo data frame
pollen = gpd.GeoDataFrame(
pollen, geometry=gpd.GeoSeries.from_wkt(pollen["wkt"]), crs="EPSG:4326"
)
# drop unnecessary columns
pollen = pollen.drop(columns=["Lat", "Long", "wkt"])
# reproject to BNG
pollen = pollen.to_crs("epsg:27700")
# get list of months with available data
monthList = list(pollen["year-month"].str[0:3].unique())
# get list of pollen monitoring sites
pollen_sites = pollen[["Site", "geometry"]].drop_duplicates()
# aggregate monthly data for each site and save as GeoPackage layers
for m in monthList:
pollen_month = pollen[pollen["year-month"].str.contains(m)]
pollen_month = pollen_month.groupby(["Site"]).mean("MaxPoaceaeConc")
pollen_month = pd.merge(pollen_month, pollen_sites, on="Site")
pollen_month.to_file("data/pollen-data.gpkg", layer=m)
Footnotes
-
Met Office. 2022. Pollen Allergies. ↩
-
Brennan, G., S. Creer, and G. Griffith. 2020. Abundance of Airborne Pollen for Nine Grass Species, Measured by qPCR, UK, 2016-2017 [Dataset] [CSV]. NERC Environmental Information Data Centre. ↩
-
Ordnance Survey (GB). 2021. OS Open Zoomstack [Dataset] [GeoPackage] [v2021-12].. ↩
-
Ordnance Survey (GB). 2021. OS Boundary-Line™ [Dataset] [GeoPackage] [v2021-10]. ↩
-
Ordnance Survey (GB). 2021. OrdnanceSurvey/OS-Open-Zoomstack-Stylesheets/GeoPackage/QGIS Stylesheets (QML)/Outdoor style/names.qml.. GitHub. ↩
-
Ordnance Survey (GB). 2021. OrdnanceSurvey/Boundary-Line-stylesheets/Geopackage stylesheets/QGIS Stylesheets (QML)/country_region.qml. GitHub. ↩
-
Rudis, B., N. Ross, and S. Garnier. 2021. Introduction to the viridis color maps. ↩
-
Waskom, M. 2021. Choosing color palettes - seaborn 0.11.2 documentation. seaborn. ↩
-
Brewer, C. and M. Harrower. 2021. ColorBrewer: Color Advice for Maps. The Pennsylvania State University. ↩